
Technical Data
技术资料
Data-independent acquisition-based SWATH-MS for quantitative proteomics: a tutorial
原作者:Christina Ludwig , Ludovic Gillet, George Rosenberger , Sabine Amon, Ben C Collins & Ruedi Aebersold
翻译:易算生物,沈诚频
DIA和SWATH-MS的历史发展和术语来源
数据非依赖采集(DIA)的基本原理是不管某个肽段的母离子是否能够检测到,采集所有能够采集的碎片离子信息。这个概念最早由Masselon等在2000年作为一个论证原理的研究发表(Masselon等,2000)。很快就有多个课题组进一步发展了这个理念并实现了同时碎裂了所有肽段(Bateman等,2002; Purvine等,2003),且利用了顺序提取母离子窗口的技术(Venable等,2004)。其后,研究者探索开发了多种不同的DIA方法,数据采集方式及分析方式 (Carvalho等,2010; Egertson等,2013; Geiger等,2010; Gillet等,2012; Panchaud等,2011; Panchaud等,2009; Plumb等,2006; Purvine等,2003; Silva等,2005; Venable等,2004; Weisbrod等,2012) 。这些方法主要在使用的质谱型号上,采集参数上和数据分析策略上有所不同。近期的一些综述比较了不同的DIA技术并讨论了各自的优缺点 (Chapman等,2014; Law & Lim,2013; Meyer & Schilling,2017; Sajic等,2015)。
“DIA”的定义最早由Venable在2004年提出(Venable等,2004),和数据依赖性采集技术 (DDA) 不同点在于DDA需要选择特定母离子进行碎裂。最初,DIA数据分析用的和DDA数据类似的搜库方法,要么是搜索混合MS2谱图(Panchaud等,2009; Venable等,2004) 或者搜索共流出的母离子信息构建的虚拟MS2谱图(Bern等,2010; Geromanos等,2009; Li等,2009)。在2012年,DIA数据分析的全新策略由Gillet提出,通过预构建谱图库来帮助进行DIA数据分析 (Gillet等,2012)。类似于靶向蛋白质组学的选择/多重反应检测方法,Gillet采用连续采集色谱流出的同一个肽段的碎片离子,并对预先采集的谱图数据库里具有可匹配的质谱、色谱参数的多肽信息进行数据挖掘来得到定性结果。作者将该方法类比为地球物理学中相关的卫星或飞行器采集地球表面图像数据的方法,命名为SWATH-MS,swaths意为这些采集的图像可以通过电脑模拟还原为地球的一整幅图像。在SWATH-MS中,“swath”意为以由质谱选定一个特定母离子隔离窗口范围,对一系列肽段质量范围内的所有离子进行碎裂。
受益于数据分析和质谱采集速度、质量精度和分辨率的显著进步,最近几年来对DIA领域的研究兴趣和研究数量大大增加(Doerr,2015)。涌现出了大量的DIA技术改进方法(图1)和多个自动化DIA数据分析工具(Bilbao等,2015)。这些进步使得蛋白质组学实验室逐渐将高质量的DIA检测纳入常规检测实验中并成功的应用于不同领域的研究中(Aebersold等,2016)。
DIA以采集和数据分析策略不同来区分,有着多种命名。不少命名是由质谱厂商申请的商标而定。本文中我们以DIA来定义这项技术,而当我们提到 “SWATH-MS”时,具体指的是由Gillet等提出的特定的采集技术和分析方法(Gillet等,2012)。

图1: DIA方法的历史发展及其对应的隔离窗口大小
自2000年初DIA技术开始兴起后,各种DIA方法的不同点在于其采集来源的质谱仪器差异、数据分析策略和特定的隔离窗口大小不同。随着仪器采集速度和分辨率的技术进步,最近几年来科学家对DIA的研究兴趣日益浓厚。多个新的中等母离子隔离窗口(4-100m/z)的技术被陆续提出。参考文献: Shotgun-CID (Purvine等,2003),Original DIA (Venable等,2004) ,MSE (Silva等,2006),PAcIFIC (Panchaud等,2009),XDIA (Carvalho等,2010),AIF (Geiger等,2010),SWATH-MS (Gillet等,2012),FT-ARM (Weisbrod等,2012),HDMSE (Geromanos等,2012),MSX (Egertson等,2013),pSMART (Prakash等,2014),WiSIM-DIA (Martin等,2016),UDMSE (Distler等,2014),HRM (Bruderer等,2015),variable window SWATH-MS (Zhang等,2015),SONAR (Moseley等,2018)。
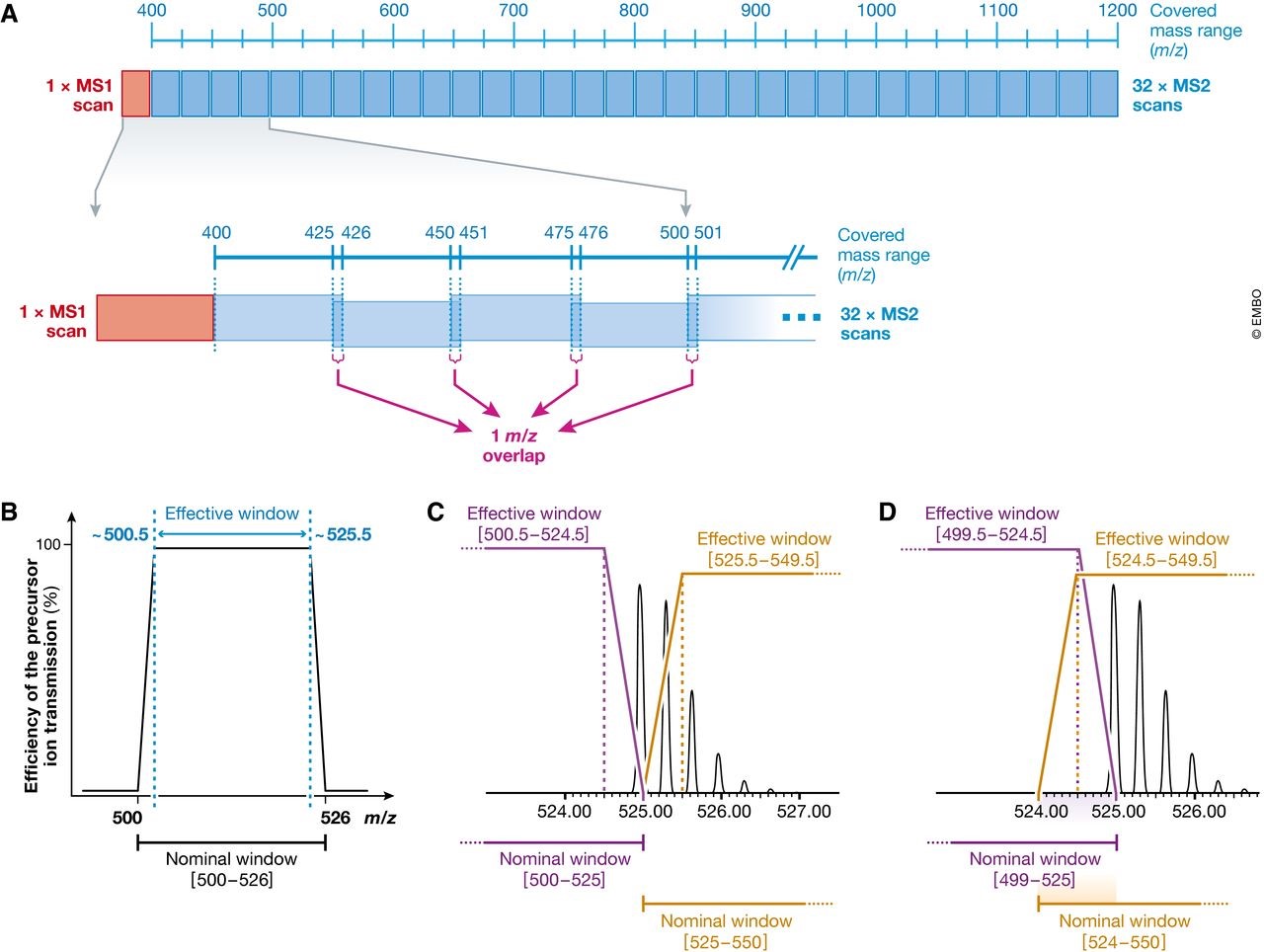
图2 使用略微重叠母离子隔离窗口的基本原理 (A) 在最早的SWATH‐MS实现方法中, 相邻SWATH窗口采用了1个m/z重叠,从而避免了隔离窗口边界上的离子传输效率下降的问题 (B) 并且将两个窗口间的母离子的同位素峰割裂效应降低 (C和D)。(B)离子传输效率问题的一个示意图,以[500–526] m/z的离子范围在目前最新的四级杆质量分析器中检测情况为例(C) Theoretical isotopic distribution of the doubly charged peptide 两电荷肽段MLSYPITIGSLLHK (m/z = 524.965)理论同位素分布情况。如果采用无重叠,其同位素峰在被分隔的同时,在两个窗口内都会落在离子传输效率低下的范围内。[有效离子传输窗口是 (500.5–524.5)和(525.5–549.5)]。 即使将窗口边缘放置到没有母离子的位置(如525.1),离子传输效率下降和同位素峰割裂的情况依然存在。(D) 重叠窗口采集[(499–525)和(524–550)]时,大部分母离子都将处于四级杆不损失传输效率的范围内,[如有效窗口为(499.5–524.5)和(524.5–549.5)]。
DIA和SWATH-MS的发展历史及相关术语
SWATH-MS基本概念
SWATH-MS检测通常依赖于质谱的一级快速扫描 (通常至少10 Hz),高分辨(通常MS2的FWHM分辨率至少15000)以及高质量精度 (至少50ppm) ,其常见配置为第一级质量分析器为四级杆 (Q)而二级质量分析器为飞行时间(TOF)或Orbitrap(图3A)。近年来在四级杆设计上的技术进步使其能够有效的在整个近乎方形的母离子隔离窗口中传输离子(图4B)。这个重要的改进保证了获取到的信号的强度,并因此被检测肽段的定量值不再会因为位于隔离窗口的不同位置而变化了。
在SWATH-MS数据采集过程中,质谱反复记录了整个色谱梯度时间内一系列MS2扫描信息。而每个采集循环中MS1扫描信息可以只记录一次或多次 (图3A)。 其中关键的一点就是所有的MS2扫描是如何进行的与这些MS1并无关系。在每个MS2扫描时,都会隔离出一个相当宽的母离子窗口。该母离子隔离窗口的m/z会逐步递增,直到完整扫描预设的整个质量范围(图3A)。为了保证足够快速的记录MS1和MS2以进行色谱峰提取和肽段定量,需要对MS2扫描数量,每个扫描的累积时间,母离子隔离窗口大小,肽段扫描质量范围等参数均需要进行优化权衡。比如,最早提出SWATH-MS的Gillet用的采集方案是32个相同的25m/z的母离子隔离窗口,覆盖了400到1200m/z的质量范围,每个扫描的累积时间是100ms (图3B)。
紧接着而来的一个重要结论就是如此大的母离子隔离窗口将会同时引入大量肽段,并将它们同时碎裂(图3C和1D)。取决于样品复杂度,这些窗口里会同时引入数十个甚至数百个共碎裂的肽段(Rost等,2012),从而产生了高度混合而卷积的MS2谱图。如此高度多路的MS2谱图无法再用传统的基于数据库匹配的DDA谱图搜索软件直接进行分析。因此SWATH-MS数据需要更先进的数据处理策略。当然,DDA和靶向质谱通常也要设置0.5到3个m/z的隔离窗口,使得其碎片谱图一样会有来自于多个母离子的卷积碎片谱峰,但毕竟这样的谱峰复杂度还是远远低于DIA的(Michalski等,2011; Wang等,2014)。解决方法之一称之为“peptide-centric scoring” (Ting等,2015)。在这项策略中,采集到的数据集会和一套已知肽段各类参数的数据集进行比对搜索。最常用的方法就是预先采集一套谱图数据库。该策略也被称之“targeted data extraction”,也是在最早发表的SWATH-MS文献中提出 (Gillet等,2012) ,而至今为止也是分析DIA数据的主要手段。
总的来说,在SWATH-MS实验中,所有落在隔离窗口范围内的离子化的多肽都将被同时碎裂并将被系统化的、无歧视的一并记录下来。SWATH-MS数据的定性和定量分析依赖于从高度多路的MS2谱图中提取,因此非常依赖于先进的数据处理策略,比如peptide-centric scoring。SWATH-MS是对追求蛋白质组全覆盖的DDA技术和追求高可重现性的靶向蛋白质组的一种补充。
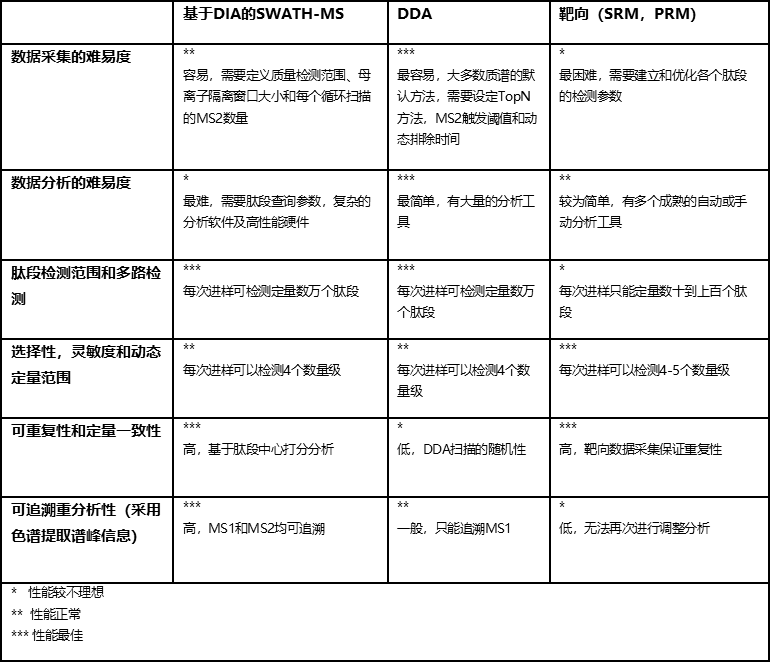
表1 优势和劣势:SWATH-MS和DDA及靶向(SRM,PRM)蛋白质组技术比较
SWATH-MS的优势
从上面总结的SWATH-MS的原理可以推导出其可能的性能优势和限制,我们在表1中进行了总结。SWATH-MS 和其他DIA技术的优势场景及其近期流行的原因均在引文中总结(Doerr,2015) ,接下来,我们也对其优势进行讨论。
数据采集的难易度
SWATH-MS的一项重要技术优势就是简化了数据采集流程。一次SWATH-MS采集需要定义母离子隔离方式(包括窗口峰宽、m/z采集范围,MS2扫描累积时间)。一旦方法确认,同一套系统中所有相似类型的样品均可采用统一的相同方法进行实验分析了。即使样品类型不同,同样的采集方法也被证明依然很可靠。这种数据采集方式的简便性和DDA采集差别不大,比靶向方法来得简单得多。像peptide precursor ions (PRM)或者peptide-fragment ion pairs (SRM)就需要预先定义每个感兴趣肽段的检测参数,且采集时经常需要确认好相应的保留时间 (Picotti & Aebersold,2012)。
肽段检测范围和多路检测
SWATH-MS的第二项优势是其对肽段定性和定量的高多路性能(一张MS2谱图含有多个肽段的碎片离子)。和基于DDA的方法类似,不同于靶向蛋白质组学,单次SWATH-MS检测可以定性超过10000种肽段。这早已在诸如人细胞系HEK293中进行了验证,通常这类样品在1%蛋白FDR水平下,2小时检测可以找30000到40000种多肽,对应于4000-5000种蛋白质 (Collins等,2017)。因此,相对于SRM或PRM方法,SWATH-MS实现了巨大的定量肽段数量提升。定量肽段越多,相应的可定量蛋白也越多,同时检测到的蛋白序列覆盖度也越高。这样可以改善蛋白组装的效果,相应带来更精确和准确的蛋白定量结果,进一步也可以帮助进行蛋白质翻译后修饰和可变剪切的检测。还有就是高多路检测率也可以提升全局数据归一化的效果。我们通常认为各个样品间大部分蛋白的浓度应该是不变的,但在SRM和PRM中,通常只检测目标肽段,因此按照这个逻辑进行样品间的全局归一化非常困难(尤其是目标肽段通常都是我们假定是参与的研究中生物学过程调控的蛋白)。SWATH-MS和DDA一样可以平行检测一系列在生物调控过程中无偏差的肽段,这是全局归一化的前提条件。
可重复性和定量一致性
当研究数百的样品的蛋白质组学时,肽段检测的可重复性及定量一致性就变成了主要困难。在DDA蛋白质组实验中要求肽段定量的高度一致性是非常困难的,因为你无法要求质谱DDA实时采集信号的时候所用的启发式母离子选择方式能够实现可重复性。因此当一个肽段在某次DDA检测中没有被定性成功,你不能判断该肽段到底是否可检出(“真阴性肽段定量”)。其实际情况很可能仅仅是因为质谱恰好因为一堆共流出肽段的存在,没有去碎裂这条多肽(“假阴性肽段定量”) (Michalski等,2011)。当然也有相应的降低这些假阴性的改进方法,比如尝试将某些样品中定性成功的肽段信息在不同样品中进行传递,从而大大改善定量数据矩阵的完整性 (Cox等,2014; Mueller等,2007; Prakash等,2006)。
相反的,在SRM,PRM和SWATH-MS中,MS2数据采集相当的有系统性且分析策略是以肽段为中心。通过这些方法的结合,我们可以较为可信的推导出目标肽段是的确“没被检测到”并因此推断出该样本中此肽段的含量至少是低于最低检测限的。尽管如此,当我们用自动化数据分析工具来检索数千种蛋白或数万种肽段时,SWATH-MS的 “假阴性肽段定量”在某种程度上依然存在。当然,和DDA一样,SWATH-MS的分析软件(Rost等,2016)也会进行样品间谱峰间对齐和鉴定结果转移。相对于母离子检测来说,碎片离子检测的灵敏度更高,且包含的信息也更丰富 (MS2中,肽段能够共流出大量碎片离子而MS1中一个肽段只能流出一个离子),因此这种结果转移方式相对于DDA来说更稳健、更灵敏。
最后一点,SRM (Abbatiello等,2015; Addona等,2009)和SWATH-MS (Collins等,2017)技术都被认为能够实现非常好的跨实验室和实验室内的结果重现性,且变异系数能够低于20% CV,即使在多个不同国家实验室进行的大规模研究也能达到这种程度。因此,如此高的重复性和一致性可以帮助生物学研究解决很多问题 (Rost等,2015)。
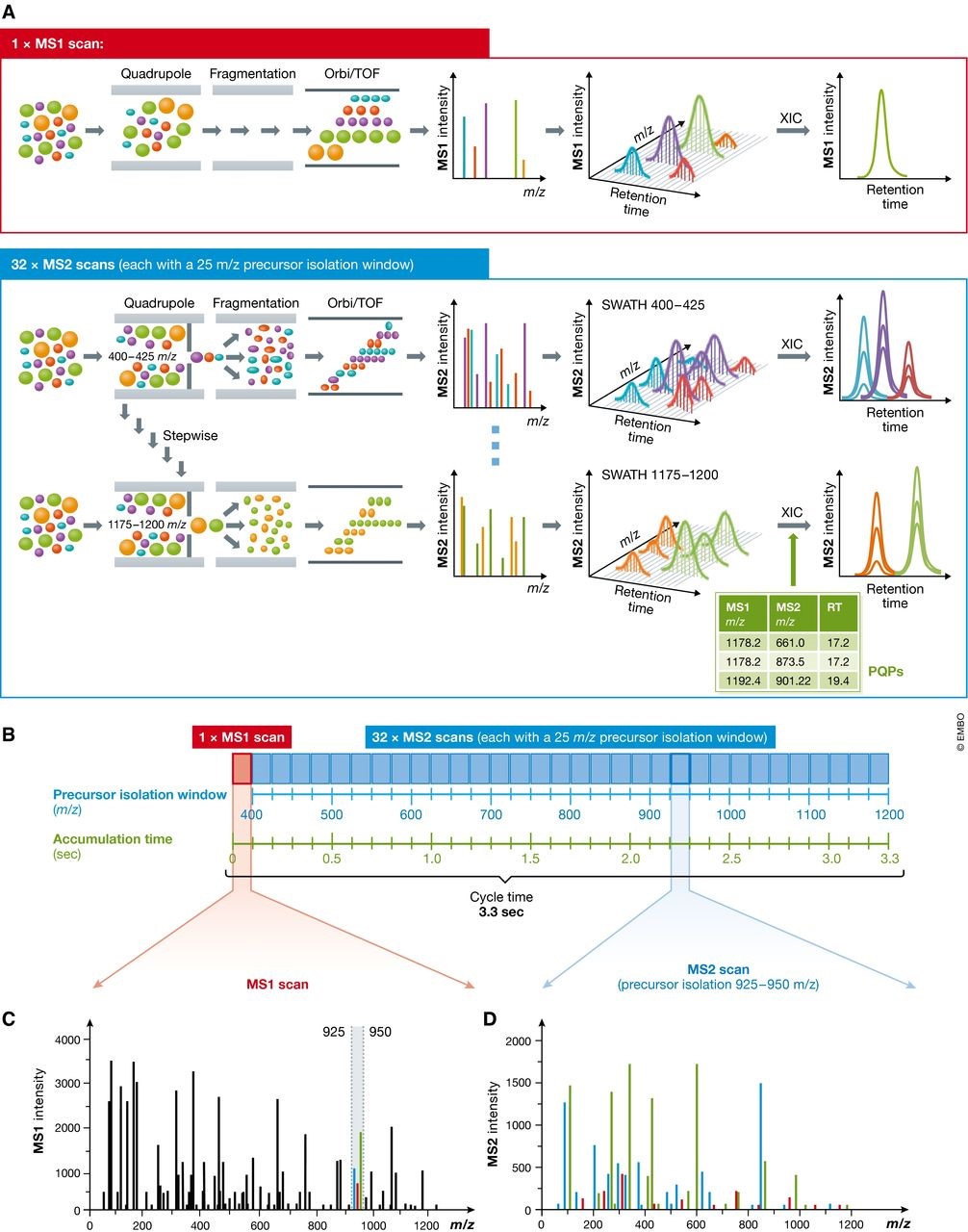
图3 SWATH-MS技术中顺序窗口数据非依赖采集的原理 (A) SWATH‐MS检测通常在具备快速质量扫描的杂交质谱上进行,主要在第一级质量分析器是四级杆而第二级质量分析器是TOF或Orbitrap的质谱上进行。在SWATH‐MS模式下,通常MS1扫描一次后会紧跟着一系列母离子隔离窗口很宽的MS2谱图(比如25 m/z的窗口大小)。通过在预先设定的质量范围内反复循环的连续母离子隔离扫描,质谱能够记录相对完整的谱图数据,其中包含了所有可检测的母离子和碎片离子的完整而连续谱峰信息。由此,MS1和MS2的提取离子色谱均可由此产生。而SWATH‐MS数据的分析则可以采用基于肽段的打分策略,需要预先记录所有可检索的肽段的色谱及质谱特征信息,从而合成一个由peptide query parameters (PQPs)构成的知识库来帮助进行肽段匹配。(B) 由Gillet et al (2012)提出的SWATH‐MS数据采集流程,在Q-TOF质谱上采集,采用预先设定的32个相同的25m/z的母离子隔离窗口,覆盖了400到1200m/z的质量范围,每个完整扫描的累积时间是100ms,完整的循环时间是3.3秒。 (C) 在某个时间点流出的MS1全扫描能够检测到的所有肽段母离子。比如质量范围在925到950 m/z,有三个共流出肽段被检测到(绿色,红色和蓝色标记)。 (D) 925–950 m/z范围内这3个肽段母离子产生的混合碎片离子MS2谱峰
可追溯查询
SWATH-MS数据的色谱和质谱的MS1母离子及MS2碎片离子谱峰信息都相当完整。这和DDA数据形成鲜明对比,DDA采集时只有母离子是连续记录而碎片信息则并不完整。SWATH-MS数据因此非常适合于过了一段时间后的追溯性分析,在最初的生物学假设中并不关注的信息或者早期无法确认的一些肽段分析参数,比如新获得的蛋白序列、肽段或者新的后修饰就可以再次进行检索分析(Rosenberger等,2017)。这不像SRM或PRM分析,如果要检测新的蛋白或肽段就必须得重新实验。
分析修饰肽段
这个优势是其对修饰肽段的定性和定量能力,它能够定位肽段序列中发生修饰的氨基酸位点,也能够搜索之前非预期的分析物。这些重要的能力是由SWATH-MS检测中对肽段整个流程时间内循环记录其高分辨、高精度MS2谱图所带来的收益。因此,不像DDA由于存在随机采集MS2和动态排除等问题,SWATH-MS可以将数据中的XIC相关MS1和MS2的信息最大化保留。还有就是对于PTM位点定位来说,碎片离子决定了修饰氨基酸的位置并且完整的MS2谱图可以用来判断不同可能位点间置信度差异(Keller等,2016; Meyer等,2017; Rosenberger等,2017)。最后一点,SWATH-MS和开放式修饰搜索工具结合可以发现非预期的全新修饰,且并不会造成搜索空间的组合爆炸 (Keller等,2016; Wang等,2015)。然后,当采用SWATH-MS来分析修饰肽段时也有和其他鸟枪法蛋白质组检测方式一样的固有挑战,亦即是修饰肽段大部分的浓度总是远远低于非修饰肽段。
总的来说,以上总结的SWATH-MS性能优势可以对复杂样本中海量的肽段进行精确定量,并能达到极高的重复性和一致性。典型的应用场景包括系统生物学研究 (Bensimon等,2012),基因相关的研究 (Liu等,2015; Okada等,2016; Williams等,2016) ,临床普筛(Liu等,2014; Sajic等,2015),药物/干扰组学普筛(Litichevskiy等,2017) 或者探索性的基础研究 (Collins等,2013; Lambert等,2013; Parker等,2015; Schubert等,2015b; Selevsek等,2015)。SWATH-MS同样适合进行快速检测分析,并能够单针检测就实现在复杂哺乳动物样本中约50%的蛋白质组覆盖率(Bruderer等,2017)
SWATH-MS的缺陷和挑战
先验知识和预构建谱图库的获取难度
在最常规且成功应用SWATH-MS的分析流程中,肽段中心打分算法依赖于关于目标肽段的色谱、质谱特性先验知识所形成的高质量谱图库 (Schubert等,2015a)。通常谱图库由DDA方式产生,由此带来的问题就是只有DDA实验中能够检测到的肽段才可能在DIA中分析到。最近几年,不依赖于谱图库的SWATH-MS 数据分析替代方案也有了长足发展,比如DIA-Umpire (Tsou等,2015),FT-ARM (Weisbrod等,2012),和PECAN (Ting等,2017)。
数据分析的难度
目前,SWATH-MS 实验分析中主要的挑战就是肽段中心打分算法部分。高度多路的MS2谱图可能由多达10个到100多个共碎裂的肽段母离子形成,因此依赖于一套精巧的分析路线来检测和定量肽段并将它们进行可靠的统计分析。对于DDA蛋白质组学来说,20多年来已经发展了丰富的分析路线 (Cox & Mann,2008; Deutsch等,2015; Keller等,2005; Reinert & Kohlbacher,2010)。而靶向蛋白质组学分析软件也已经成熟应用了好几年了(Colangelo等,2013; Malmstrom等,2012; Teleman等,2012)。相应的自动化分析SWATH-MS肽段谱的软件还处于发展阶段,其中一些也已经得到了广泛应用并供免费使用(相应的综述参考 (Bilbao等,2015))。对其中五种广泛使用的软件的综合比较文献可以参考(Navarro等,2016)。
选择性,灵敏度和动态定量范围
SWATH-MS还有一个主要的挑战是其定量的态范围依然有限。多个课题组报道了在大规模复杂蛋白质组样本中碎片离子(MS2)具有比相应母离子(MS1)更高的信噪比,选择性和同一个扫描内的动态范围(Egertson等,2013; Gillet等,2012; Venable等,2004)。在复杂样本中,肽段的MS1信号很有可能被共流出的非常接近或相同m/z的信号所干扰。而在MS2水平的信号则仅来自于MS1隔离窗口内的这些母离子信号碎片,而由于MS2定量会选择多个离子碎片,其均受到干扰的可能性非常之低。另外考虑到离子阱质谱的选择性能力,MS1扫描的信号受到其AGC(auto gain control)效应的影响,高丰度肽段信号会抑制低丰度肽段,而SWATH-MS中由于只检测隔离窗口内的母离子碎片,该效应得到很好地改善。目前SWATH-MS在Q-TOF仪器上使用时,可以在单针检测复杂的注入人细胞系全酶解液的实验中覆盖4到4.5个数量级定量动态范围(Collins等,2017)。文献报道中的复杂样品定量最低检测限大约为中等attomole到低femtomole(上柱量)。如此定量范围还是比最新的SRM或MRM检测的灵敏度低3-10倍 (Gillet等,2012; Liu等,2013; Schmidlin等,2016) 。
参考文献
1. Abbatiello SE, Schilling B, Mani DR, Zimmerman LJ, Hall SC, MacLean B, Albertolle M, Allen S, Burgess M, Cusack MP, Gosh M, Hedrick V, Held JM, Inerowicz HD, Jackson A, Keshishian H, Kinsinger CR, Lyssand J, Makowski L, Mesri M et al. (2015) Large-Scale Interlaboratory Study to Develop, Analytically Validate and Apply Highly Multiplexed, Quantitative Peptide Assays to Measure Cancer-Relevant Proteins in Plasma. Molecular & cellular proteomics : MCP 14: 2357-2374
2. Addona TA, Abbatiello SE, Schilling B, Skates SJ, Mani DR, Bunk DM, Spiegelman CH, Zimmerman LJ, Ham AJ, Keshishian H, Hall SC, Allen S, Blackman RK, Borchers CH, Buck C, Cardasis HL, Cusack MP, Dodder NG, Gibson BW, Held JM et al. (2009) Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nature biotechnology 27: 633-641
3. Aebersold R, Bensimon A, Collins BC, Ludwig C, Sabido E (2016) Applications and Developments in Targeted Proteomics: From SRM to DIA/SWATH. Proteomics 16: 2065-2067
4. Bateman RH, Carruthers R, Hoyes JB, Jones C, Langridge JI, Millar A, Vissers JP (2002) A novel precursor ion discovery method on a hybrid quadrupole orthogonal acceleration time-offlight (Q-TOF) mass spectrometer for studying protein phosphorylation. Journal of the American Society for Mass Spectrometry 13: 792-803
5. Bensimon A, Heck AJ, Aebersold R (2012) Mass spectrometry-based proteomics and network biology. Annual review of biochemistry 81: 379-405
6. Bern M, Finney G, Hoopmann MR, Merrihew G, Toth MJ, MacCoss MJ (2010) Deconvolution of mixture spectra from ion-trap data-independent-acquisition tandem mass spectrometry. Analytical chemistry 82: 833-841
7. Bilbao A, Varesio E, Luban J, Strambio-De-Castillia C, Hopfgartner G, Muller M, Lisacek F (2015) Processing strategies and software solutions for data-independent acquisition in mass spectrometry. Proteomics 15: 964-980
8. Bruderer R, Bernhardt OM, Gandhi T, Miladinovic SM, Cheng LY, Messner S, Ehrenberger T, Zanotelli V, Butscheid Y, Escher C, Vitek O, Rinner O, Reiter L (2015) Extending the limits of quantitative proteome profiling with data-independent acquisition and application to acetaminophen-treated three-dimensional liver microtissues. Molecular & cellular proteomics : MCP 14: 1400-1410
9. Bruderer R, Bernhardt OM, Gandhi T, Xuan Y, Sondermann J, Schmidt M, Gomez-Varela D, Reiter L (2017) Optimization of Experimental Parameters in Data-Independent Mass Spectrometry Significantly Increases Depth and Reproducibility of Results. Molecular & cellular proteomics : MCP
10. Carvalho PC, Han X, Xu T, Cociorva D, Carvalho Mda G, Barbosa VC, Yates JR, 3rd (2010) XDIA: improving on the label-free data-independent analysis. Bioinformatics (Oxford, England) 26: 847-848
11. Chapman JD, Goodlett DR, Masselon CD (2014) Multiplexed and data-independent tandem mass spectrometry for global proteome profiling. Mass spectrometry reviews 33: 452-470
12. Colangelo CM, Chung L, Bruce C, Cheung KH (2013) Review of software tools for design and analysis of large scale MRM proteomic datasets. Methods (San Diego, Calif) 61: 287-298
13. Collins BC, Gillet LC, Rosenberger G, Rost HL, Vichalkovski A, Gstaiger M, Aebersold R (2013) Quantifying protein interaction dynamics by SWATH mass spectrometry: application to the 14-3-3 system. Nature methods 10: 1246-1253
14. Collins BC, Hunter CL, Liu Y, Schilling B, Rosenberger G, Bader SL, Chan DW, Gibson BW, Gingras AC, Held JM, Hirayama-Kurogi M, Hou G, Krisp C, Larsen B, Lin L, Liu S, Molloy MP, Moritz RL, Ohtsuki S, Schlapbach R et al. (2017) Multi-laboratory assessment of reproducibility, qualitative and quantitative performance of SWATH-mass spectrometry. Nature communications 8: 291
15. Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M (2014) Accurate proteome-wide labelfree quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Molecular & cellular proteomics : MCP 13: 2513-2526
16. Cox J, Mann M (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature biotechnology 26: 1367-1372
17. Deutsch EW, Mendoza L, Shteynberg D, Slagel J, Sun Z, Moritz RL (2015) Trans-Proteomic Pipeline, a standardized data processing pipeline for large-scale reproducible proteomics informatics. Proteomics Clinical applications 9: 745-754
18. Distler U, Kuharev J, Navarro P, Levin Y, Schild H, Tenzer S (2014) Drift time-specific collision energies enable deep-coverage data-independent acquisition proteomics. Nature methods 11: 167-170
19. Doerr A (2015) DIA mass spectrometry. Nat Meth 12: 35-35
20. Egertson JD, Kuehn A, Merrihew GE, Bateman NW, MacLean BX, Ting YS, Canterbury JD, Marsh DM, Kellmann M, Zabrouskov V, Wu CC, MacCoss MJ (2013) Multiplexed MS/MS for improved data-independent acquisition. Nature methods 10: 744-746
21. Geiger T, Cox J, Mann M (2010) Proteomics on an Orbitrap benchtop mass spectrometer using all-ion fragmentation. Molecular & cellular proteomics : MCP 9: 2252-2261
22. Geromanos SJ, Hughes C, Ciavarini S, Vissers JP, Langridge JI (2012) Using ion purity scores for enhancing quantitative accuracy and precision in complex proteomics samples. Analytical and bioanalytical chemistry 404: 1127-1139
23. Geromanos SJ, Vissers JP, Silva JC, Dorschel CA, Li GZ, Gorenstein MV, Bateman RH, Langridge JI (2009) The detection, correlation, and comparison of peptide precursor and product ions from data independent LC-MS with data dependant LC-MS/MS. Proteomics 9: 1683-1695
24. Gillet LC, Navarro P, Tate S, Rost H, Selevsek N, Reiter L, Bonner R, Aebersold R (2012) Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Molecular & cellular proteomics : MCP 11: O111.016717
25. Keller A, Bader SL, Kusebauch U, Shteynberg D, Hood L, Moritz RL (2016) Opening a SWATH Window on Posttranslational Modifications: Automated Pursuit of Modified Peptides. Molecular & cellular proteomics : MCP 15: 1151-1163
26. Keller A, Eng J, Zhang N, Li XJ, Aebersold R (2005) A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Molecular systems biology 1: 2005.0017
27. Lambert JP, Ivosev G, Couzens AL, Larsen B, Taipale M, Lin ZY, Zhong Q, Lindquist S, Vidal M, Aebersold R, Pawson T, Bonner R, Tate S, Gingras AC (2013) Mapping differential interactomes by affinity purification coupled with data-independent mass spectrometry acquisition. Nature methods 10: 1239-1245
28. Law KP, Lim YP (2013) Recent advances in mass spectrometry: data independent analysis and hyper reaction monitoring. Expert review of proteomics 10: 551-566
29. Li GZ, Vissers JP, Silva JC, Golick D, Gorenstein MV, Geromanos SJ (2009) Database searching and accounting of multiplexed precursor and product ion spectra from the data independent analysis of simple and complex peptide mixtures. Proteomics 9: 1696-1719
30. Litichevskiy L, Peckner R, Abelin JG, Asiedu JK, Creech AL, Davis JF, Davison D, Dunning CM, Egertson JD, Egri S, Gould J, Ko T, Johnson SA, Lahr DL, Lam D, Liu Z, Lyons NJ, Lu X, MacLean BX, Mungenast AE et al. (2017) A Library of Phosphoproteomic and Chromatin Signatures for Characterizing Cellular Responses to Drug Perturbations. bioRxiv
31. Liu Y, Buil A, Collins BC, Gillet LC, Blum LC, Cheng LY, Vitek O, Mouritsen J, Lachance G, Spector TD, Dermitzakis ET, Aebersold R (2015) Quantitative variability of 342 plasma proteins in a human twin population. Molecular systems biology 11: 786
32. Liu Y, Chen J, Sethi A, Li QK, Chen L, Collins B, Gillet LC, Wollscheid B, Zhang H, Aebersold R (2014) Glycoproteomic analysis of prostate cancer tissues by SWATH mass spectrometry discovers N-acylethanolamine acid amidase and protein tyrosine kinase 7 as signatures for tumor aggressiveness. Molecular & cellular proteomics : MCP 13: 1753-1768
33. Liu Y, Huttenhain R, Surinova S, Gillet LC, Mouritsen J, Brunner R, Navarro P, Aebersold R (2013) Quantitative measurements of N-linked glycoproteins in human plasma by SWATHMS. Proteomics 13: 1247-1256
34. Malmstrom L, Malmstrom J, Selevsek N, Rosenberger G, Aebersold R (2012) Automated workflow for large-scale selected reaction monitoring experiments. Journal of proteome research 11: 1644-1653
35. Martin LB, Sherwood RW, Nicklay JJ, Yang Y, Muratore-Schroeder TL, Anderson ET, Thannhauser TW, Rose JK, Zhang S (2016) Application of wide selected-ion monitoring data- independent acquisition to identify tomato fruit proteins regulated by the CUTIN DEFICIENT2 transcription factor. Proteomics 16: 2081-2094
36. Masselon C, Anderson GA, Harkewicz R, Bruce JE, Pasa-Tolic L, Smith RD (2000) Accurate mass multiplexed tandem mass spectrometry for high-throughput polypeptide identification from mixtures. Analytical chemistry 72: 1918-1924
37. Meyer JG, Mukkamalla S, Steen H, Nesvizhskii AI, Gibson BW, Schilling B (2017) PIQED: automated identification and quantification of protein modifications from DIA-MS data. Nature methods 14: 646-647
38. Meyer JG, Schilling B (2017) Clinical applications of quantitative proteomics using targeted and untargeted data-independent acquisition techniques. Expert review of proteomics 14: 419-429
39. Michalski A, Cox J, Mann M (2011) More than 100, 000 detectable peptide species elute in single shotgun proteomics runs but the majority is inaccessible to data-dependent LCMS/MS. Journal of proteome research 10: 1785-1793
40. Moseley MA, Hughes CJ, Juvvadi PR, Soderblom EJ, Lennon S, Perkins SR, Thompson JW, Steinbach WJ, Geromanos SJ, Wildgoose J, Langridge JI, Richardson K, Vissers JPC (2018) Scanning Quadrupole Data-Independent Acquisition, Part A: Qualitative and Quantitative Characterization. Journal of proteome research 17: 770-779
41. Mueller LN, Rinner O, Schmidt A, Letarte S, Bodenmiller B, Brusniak MY, Vitek O, Aebersold R, Muller M (2007) SuperHirn – a novel tool for high resolution LC-MS-based peptide/protein profiling. Proteomics 7: 3470-3480
42. Navarro P, Kuharev J, Gillet LC, Bernhardt OM, MacLean B, Rost HL, Tate SA, Tsou CC, Reiter L, Distler U, Rosenberger G, Perez-Riverol Y, Nesvizhskii AI, Aebersold R, Tenzer S (2016) A multicenter study benchmarks software tools for label-free proteome quantification. Nature biotechnology
43. Okada H, Ebhardt HA, Vonesch SC, Aebersold R, Hafen E (2016) Proteome-wide association studies identify biochemical modules associated with a wing-size phenotype in Drosophila melanogaster. Nature communications 7: 12649
44. Panchaud A, Jung S, Shaffer SA, Aitchison JD, Goodlett DR (2011) Faster, quantitative, and accurate precursor acquisition independent from ion count. Analytical chemistry 83: 2250- 2257
45. Panchaud A, Scherl A, Shaffer SA, von Haller PD, Kulasekara HD, Miller SI, Goodlett DR (2009) Precursor acquisition independent from ion count: how to dive deeper into the proteomics ocean. Analytical chemistry 81: 6481-6488
46. Parker BL, Yang G, Humphrey SJ, Chaudhuri R, Ma X, Peterman S, James DE (2015) Targeted phosphoproteomics of insulin signaling using data-independent acquisition mass spectrometry. Science signaling 8: rs6
47. Picotti P, Aebersold R (2012) Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nature methods 9: 555-566
48. Plumb RS, Johnson KA, Rainville P, Smith BW, Wilson ID, Castro-Perez JM, Nicholson JK (2006) UPLC/MS(E); a new approach for generating molecular fragment information for biomarker structure elucidation. Rapid communications in mass spectrometry : RCM 20: 1989-1994
49. Prakash A, Mallick P, Whiteaker J, Zhang H, Paulovich A, Flory M, Lee H, Aebersold R, Schwikowski B (2006) Signal maps for mass spectrometry-based comparative proteomics. Molecular & cellular proteomics : MCP 5: 423-432
50. Prakash A, Peterman S, Ahmad S, Sarracino D, Frewen B, Vogelsang M, Byram G, Krastins B, Vadali G, Lopez M (2014) Hybrid data acquisition and processing strategies with increased throughput and selectivity: pSMART analysis for global qualitative and quantitative analysis. Journal of proteome research 13: 5415-5430
51. Purvine S, Eppel JT, Yi EC, Goodlett DR (2003) Shotgun collision-induced dissociation of peptides using a time of flight mass analyzer. Proteomics 3: 847-850
52. Reinert K, Kohlbacher O (2010) OpenMS and TOPP: open source software for LC-MS data analysis. Methods in molecular biology (Clifton, NJ) 604: 201-211
53. Rosenberger G, Liu Y, Rost HL, Ludwig C, Buil A, Bensimon A, Soste M, Spector TD, Dermitzakis ET, Collins BC, Malmstrom L, Aebersold R (2017) Inference and quantification of peptidoforms in large sample cohorts by SWATH-MS. Nature biotechnology 35: 781-788
54. Rost H, Malmstrom L, Aebersold R (2012) A computational tool to detect and avoid redundancy in selected reaction monitoring. Molecular & cellular proteomics : MCP 11: 540- 549
55. Rost HL, Liu Y, D’Agostino G, Zanella M, Navarro P, Rosenberger G, Collins BC, Gillet L, Testa G, Malmstrom L, Aebersold R (2016) TRIC: an automated alignment strategy for reproducible protein quantification in targeted proteomics. Nature methods 13: 777-783
56. Rost HL, Malmstrom L, Aebersold R (2015) Reproducible quantitative proteotype data matrices for systems biology. Molecular biology of the cell 26: 3926-3931
57. Sajic T, Liu Y, Aebersold R (2015) Using data-independent, high-resolution mass spectrometry in protein biomarker research: perspectives and clinical applications. Proteomics Clinical applications 9: 307-321
58. Schmidlin T, Garrigues L, Lane CS, Mulder TC, van Doorn S, Post H, de Graaf EL, Lemeer S, Heck AJ, Altelaar AF (2016) Assessment of SRM, MRM and DIA for the targeted analysis of phosphorylation dynamics in non-small cell lung cancer. Proteomics
59. Schubert OT, Gillet LC, Collins BC, Navarro P, Rosenberger G, Wolski WE, Lam H, Amodei D, Mallick P, MacLean B, Aebersold R (2015a) Building high-quality assay libraries for targeted analysis of SWATH MS data. Nature protocols 10: 426-441
60. Schubert OT, Ludwig C, Kogadeeva M, Zimmermann M, Rosenberger G, Gengenbacher M, Gillet LC, Collins BC, Rost HL, Kaufmann SH, Sauer U, Aebersold R (2015b) Absolute Proteome Composition and Dynamics during Dormancy and Resuscitation of Mycobacterium tuberculosis. Cell host & microbe 18: 96-108
61. Selevsek N, Chang CY, Gillet LC, Navarro P, Bernhardt OM, Reiter L, Cheng LY, Vitek O, Aebersold R (2015) Reproducible and consistent quantification of the Saccharomyces cerevisiae proteome by SWATH-mass spectrometry. Molecular & cellular proteomics : MCP 14: 739-749
62. Silva JC, Denny R, Dorschel CA, Gorenstein M, Kass IJ, Li GZ, McKenna T, Nold MJ, Richardson K, Young P, Geromanos S (2005) Quantitative proteomic analysis by accurate mass retention time pairs. Analytical chemistry 77: 2187-2200
63. Silva JC, Gorenstein MV, Li GZ, Vissers JP, Geromanos SJ (2006) Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Molecular & cellular proteomics : MCP 5: 144-156
64. Teleman J, Karlsson C, Waldemarson S, Hansson K, James P, Malmstrom J, Levander F (2012) Automated selected reaction monitoring software for accurate label-free protein quantification. Journal of proteome research 11: 3766-3773
65. Ting YS, Egertson JD, Bollinger JG, Searle BC, Payne SH, Noble WS, MacCoss MJ (2017) PECAN: library-free peptide detection for data-independent acquisition tandem mass spectrometry data. Nature methods 14: 903-908
66. Ting YS, Egertson JD, Payne SH, Kim S, MacLean B, Kall L, Aebersold R, Smith RD, Noble WS, MacCoss MJ (2015) Peptide-Centric Proteome Analysis: An Alternative Strategy for the Analysis of Tandem Mass Spectrometry Data. Molecular & cellular proteomics : MCP 14: 2301-2307
67. Tsou CC, Avtonomov D, Larsen B, Tucholska M, Choi H, Gingras AC, Nesvizhskii AI (2015) DIAUmpire: comprehensive computational framework for data-independent acquisition proteomics. Nature methods 12: 258-264, 257 p following 264
68. Venable JD, Dong MQ, Wohlschlegel J, Dillin A, Yates JR (2004) Automated approach for quantitative analysis of complex peptide mixtures from tandem mass spectra. Nature methods 1: 39-45 Wang J, Bourne PE, Bandeira N (2014) MixGF: spectral probabilities for mixture spectra from more than one peptide. Molecular & cellular proteomics : MCP 13: 3688-3697
69. Wang J, Tucholska M, Knight JD, Lambert JP, Tate S, Larsen B, Gingras AC, Bandeira N (2015) MSPLIT-DIA: sensitive peptide identification for data-independent acquisition. Nature methods 12: 1106-1108
70. Weisbrod CR, Eng JK, Hoopmann MR, Baker T, Bruce JE (2012) Accurate peptide fragment mass analysis: multiplexed peptide identification and quantification. Journal of proteome research 11: 1621-1632
71. Williams EG, Wu Y, Jha P, Dubuis S, Blattmann P, Argmann CA, Houten SM, Amariuta T, Wolski W, Zamboni N, Aebersold R, Auwerx J (2016) Systems proteomics of liver mitochondria function. Science 352
72. Zhang Y, Bilbao A, Bruderer T, Luban J, Strambio-De-Castillia C, Lisacek F, Hopfgartner G, Varesio E (2015) The Use of Variable Q1 Isolation Windows Improves Selectivity in LCSWATH-MS Acquisition. Journal of proteome research 14: 4359-4371
